Egg Activation
Much of the research in our laboratory is focused on elucidating the molecular mechanisms of how oocytes and eggs commit to re-entry into the cell cycle during maturation and fertilization. Our interest in this is two-fold. First, egg activation represents an unique opportunity to study an "all-or-none" (digital) signaling response with ramifications for understanding cellular signaling in general, particularly in how it relates to cell cycle entry (with obvious ramifications for oncogenesis). Second, fertilization and egg activation represent a fundamental developmental stage-- the dramatic activation of the egg at fertilization is a one-time event in the life of an organism and is crucial to normal development. Our basic approach is to dissect the signaling pathways leading from either hormone or sperm binding at the egg surface to various outputs that represent cellular commitment steps. Our model systems have been the echinoderms – sea urchin and sea stars – and the ascidians. Both large-scale biochemical approaches as well as single cell (microinjection and microscopic imaging) experiments can be conducted easily using the large, optically clear eggs of these marine invertebrates, which share the basic aspects of egg activation with other species, including mammals.
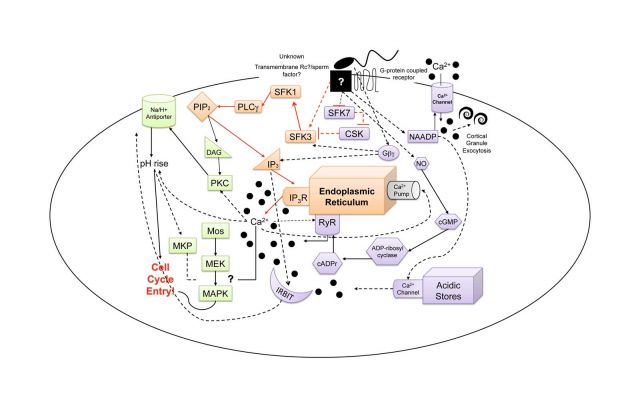
Our current focus is on the molecular mechanism of egg activation and in particular, the elucidation of the signaling components that lead to the initiation of Ca2+ release from internal egg stores at fertilization. This Ca2+ serves as the initial trigger for all subsequent egg activation events (cytoskeletal remodeling, metabolic activation, initiation of protein synthesis, re-entry into the cell cycle, etc…). In both sea star and sea urchin eggs, Src-family tyrosine kinases (SFKs) are sequentially activated and subsequently activate PLC-g, generating IP3 and thus initiating Ca2+ release through IP3 receptors in the egg’s endoplasmic reticulum (see the purple colored module in the accompanying figure). At the egg plasma membrane, it appears that there are specialized membrane microdomains that serve to scaffold and organize various signaling components including the SFKs and PLCg; these regions may aid in forming a sort of “fertilization signaling synapse” where the sperm and egg meet. To date, the echinoderms serve as one of the best model species in which even a portion of the egg activation signaling pathway is known in detail.
While the Src-PLCg pathway of calcium release is required to initiate calcium release in echinoderm eggs, there are clearly many other pathways that collectively modulate calcium in eggs (we refer to this as the “calcium tapestry” as illustrated in the accompanying figure). Our work on the mechanism of calcium release serves as a platform for several roads of investigation. Currently, we are characterizing the regulation of the MAP kinase signaling pathway and attempting to identify targets that modulate the first round of DNA synthesis (the orange module in the accompany calcium tapestry figure).
The elucidation of the California purple sea urchin (Strongylocentrotus purpuratus) genome sequence in 2006 allowed us to characterize the sea urchin egg phosphoproteome, revealing a landscape of proteins that undergo a change in phosphorylation status in response to fertilization. Our current work, in collaboration with Andrew Emili’s lab at the University of Toronto, is making use of extremely high throughput work flow platforms to quantitate phosphorylation and create network maps of more than 5000 unique phosphorylation events that occur in the first two minutes post sperm addition. The exquisite synchronicity coupled with the abundance of material offered by the sea urchin system makes this richly informative, unbiased approach possible. In essence, we are asking the egg to show us what changes coincide with the fertilization event and then we pursue promising candidates through hypothesis-driven experimental analyses.